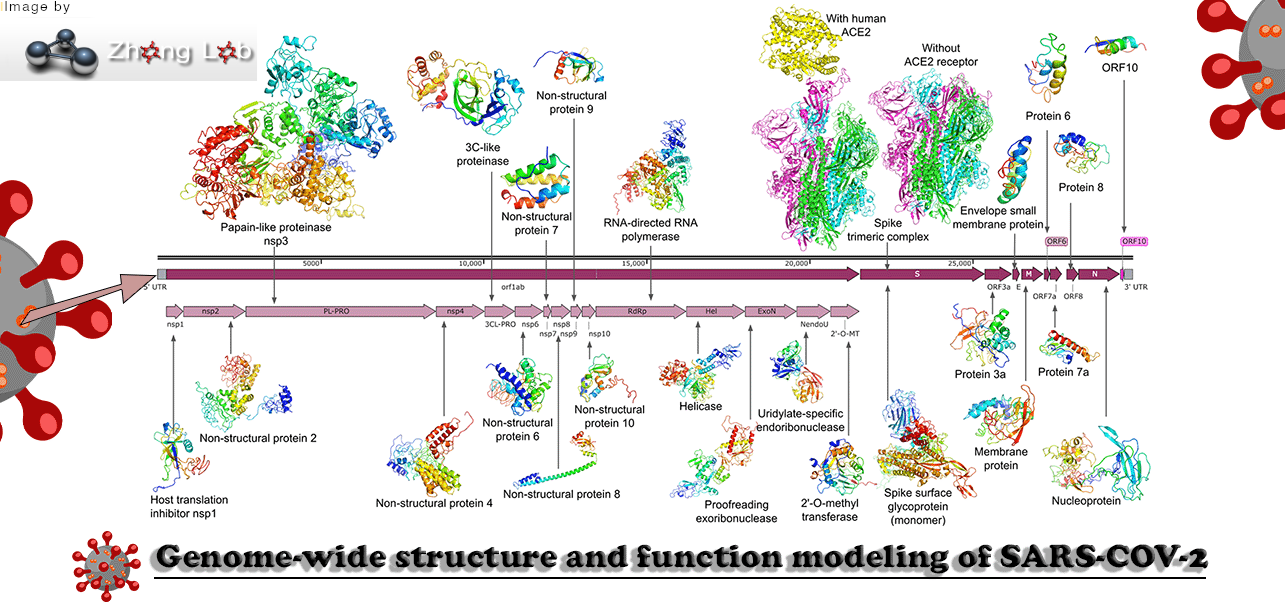
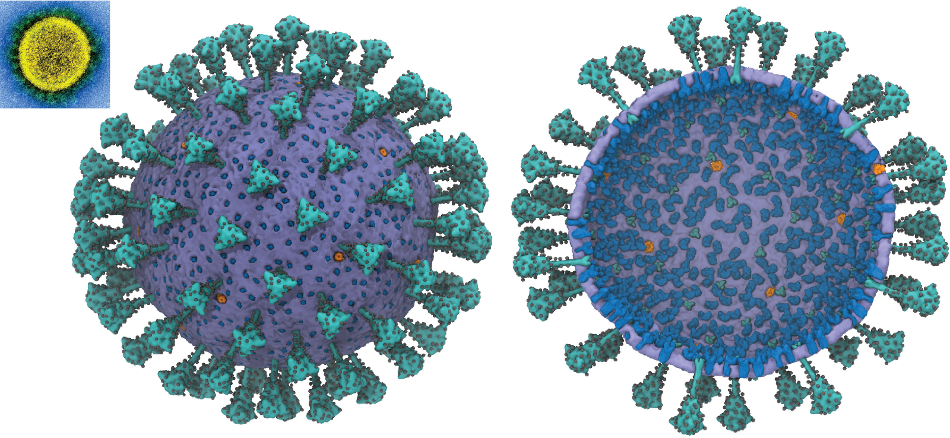
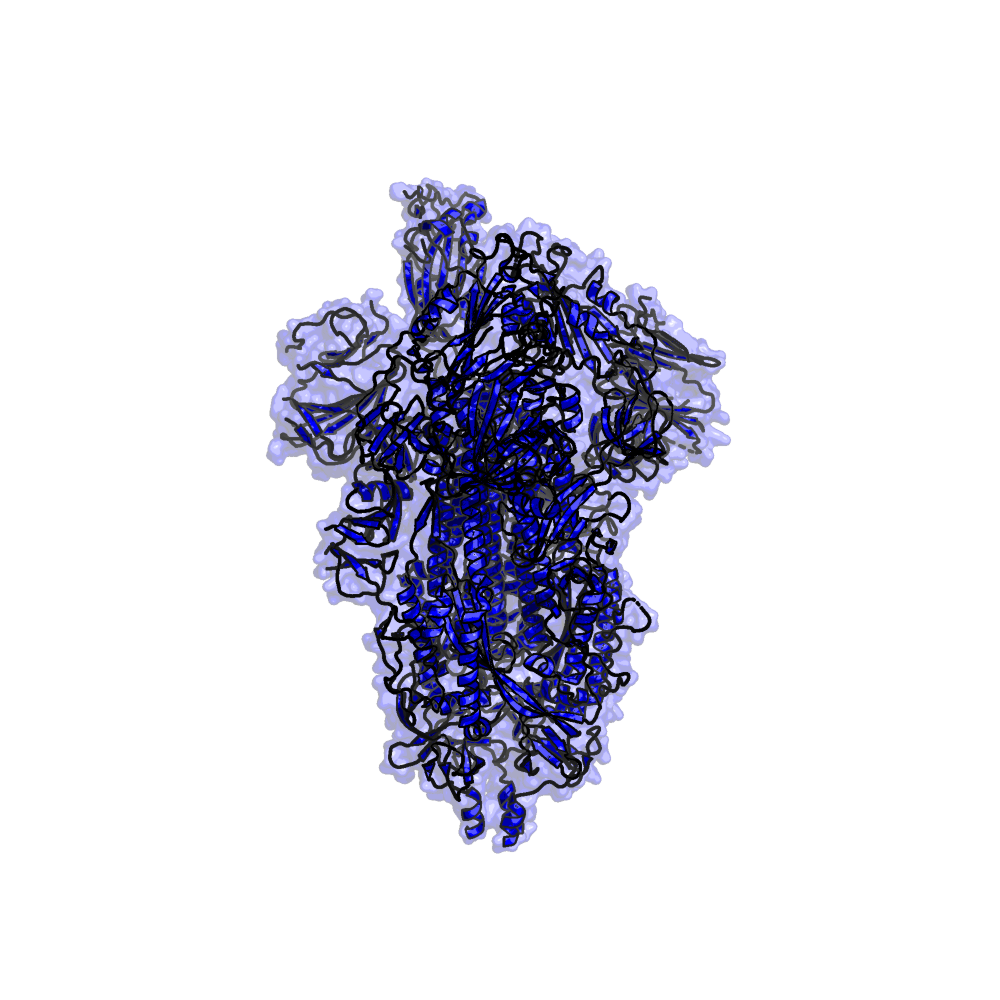
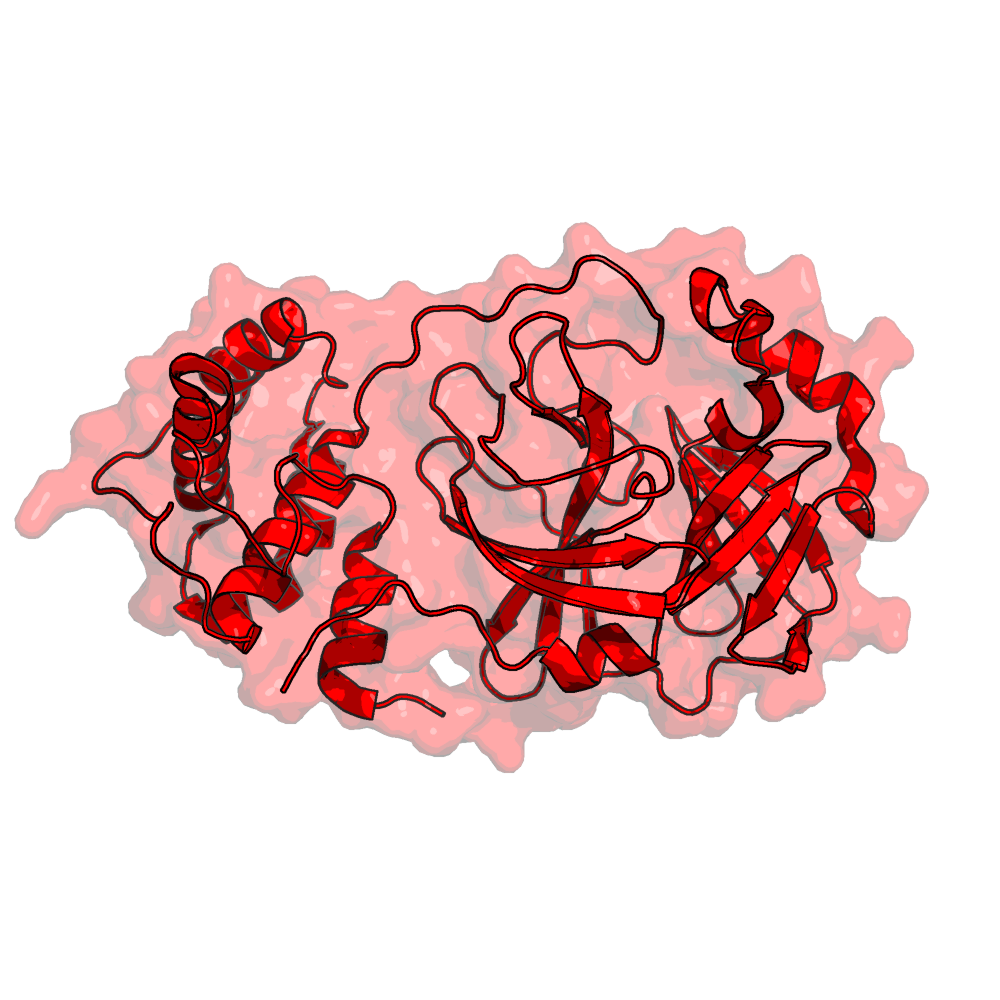
Proteins and Virion
Viral/Host Proteins and the Virion itself
Virus (SARS-CoV-2 and SARS-CoV), and Host based proteins are referenced here as well as the whole virion particle.
Data classification:- Structures: Data defining structures determined by experimental methods and referenced via a unique identifier such as a PDB ID.
- Models: Derived, integrated, or refined structures from multiple data sources prepared for different computational tasks.
- Simulations: The datasets produced as a result of applying the models to different scientific techniques.
- Unpublished data with no preprints have no indicator
- Data in a preprint or submitted for publication are given this
 marker. If the preprint is available, it will always show and should work as a link
marker. If the preprint is available, it will always show and should work as a link - Data published in a paper or accepted are given this
 marker (i.e. has been approved by formal peer review) and should work as links.
marker (i.e. has been approved by formal peer review) and should work as links.